How do you translate more than eleven-million lines of XML into modern, elegant, web pages? The Finding Aids Redesign (FADESIGN) team had to answer this question as part of a multi-year effort to replace a 20-year-old web publishing process and modernize the design of the finding aids hosted by NYU Libraries. In order to develop a solution, the team had to better understand the input.
The eleven-million lines of XML input into the finding aids publishing pipeline are not, thankfully, all in one file. Instead, the XML is distributed across more than five-thousand individual Encoded Archival Description (EAD) files originating from seven organizations housing nine different archival repositories. These XML files conform to the EAD 2002 XML schema, which is incredibly flexible and supports a wide variety of archival-description styles. This flexibility is an asset when describing archival content, but becomes an impediment when developing data structures and web page layouts used to transform the EAD data into HTML. For example, per the EAD 2002 schema, the component element (<c>) can be nested ad infinitum. How do you design a web page that handles potentially infinite nesting? Additionally, certain elements like runner (<runner>) and imprint (<imprint>) are defined in the schema but are not used by our archival repositories. Therefore, we needed to find a middle ground between the flexibility supported by the schema and the practical requirements of day-to-day archival description. Enter the data model.
The data model is a document collaboratively developed by the archivists and the development team that specifies the subset of EAD 2002 elements and attributes that we would need to faithfully represent the archival description in NYU’s finding aids. The initial data model was developed based on known practices and reviewing data types programmatically generated by the Zek tool, and refined by moving EADs through our nascent publication pipeline: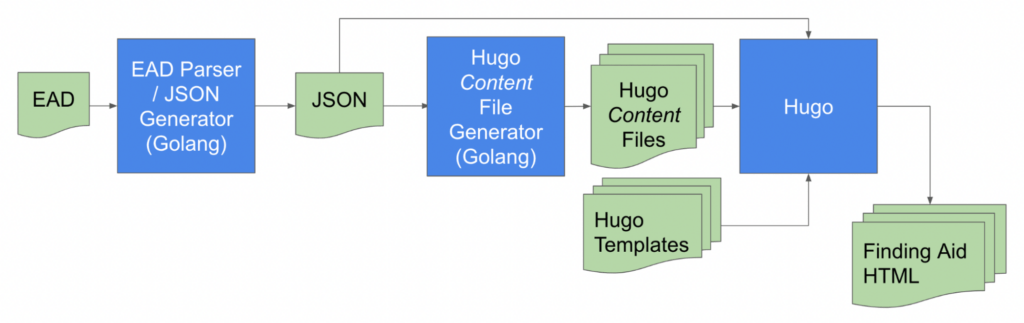
The pipeline consists of a Golang-based EAD exporter that collects EADs from ArchivesSpace instances, another Golang application that parses the incoming EAD files and generates JSON files. These JSON files and a set of Hugo template files are then input into the Hugo static site generator application to create the finding aid web pages. In order to converge on a publishing solution that met stakeholder requirements, we needed to iterate over the steps below:
- write/update Golang data types based on the data model
- parse a sample set of EADs and output JSON
- feed the JSON and Hugo templates to the Hugo static site generator application to generate finding aids
- review the finding aids, note where changes are required to the design and the data model
- update the data model
Although the above steps were executed over increasingly large sample sets of EADs, we still had concerns that, given the flexibility of the EAD 2002 schema, there were valid use cases that were not reflected in the data model. We needed more information about the structure and variation of schema elements in use. Therefore, we developed a set of tools to perform analysis on the entire set of more than 5,000 EADs.
The EAD Analysis Tools are a set of minimal-functionality scripts we created to answer the following questions:
- How many lines are in the largest EAD?
- What is the maximum nesting level of each element?
- For every element of interest:
- Does the EAD structure vary widely across archival repositories?
- Across all EADs, what sequences of child elements are present?
- What is the set of <dao @xlink:role> values across all EADs?
- Are there EADs with <dao @xlink:role> values that are not in the controlled vocabulary?
The sections below will walk you through the scripts we used to answer each of the above questions. We wrote the scripts in Ruby and Bash so that we could prototype rapidly and, in the case of the Ruby scripts, take advantage of the Nokogiri XML Ruby Gem.
Conclusion:
Understanding the input to the finding aids publication pipeline was critical to the project’s success. We developed tools that helped us refine the data model, design appropriate data structures, identify test candidates for various scenarios, gather insight into how the performance of certain operations corresponded to the EAD structure, and identify critical parameters required for the web page design. The data we gathered gave us confidence that when we went to production we were not going to be surprised by a radically different set of EADs incompatible with our publishing infrastructure. Going forward, we can use these same analysis tools to compare EADs from new archival repositories against the data model, allowing us to catch potential issues before publication.
Question: How many lines are in the largest EAD?
Motivation:
We wanted an answer to this question so that we could test our pipeline scalability and throughput. It turns out that this question is rather straightforward to answer using Bash commands and doesn’t require a stand-alone script.
Script: ad hoc
The following commands will pretty-print the EADs and then will count the number of lines in the pretty-printed files. We needed to pretty print the files because XML does not require line breaks, which means that a massive, properly formatted EAD may only contain one very long line. After pretty-printing, however, the EAD line counts can be accurately assessed. For example, without pretty printing, the file in our corpus with the greatest number of lines appears to be fales/mss_130.xml with 25,986 lines. After pretty printing, however, we find that the file with the greatest number of lines is actually cbh/bcms_0002.xml with 281,168 lines.
Please note that the pretty-print command below overwrites the original EADs with the pretty-printing version. This isn’t a problem for us because we store the EADs in a Git repository which allows us to easily restore the EADs to their original state.
Sample Execution:
# pretty-print all XML files in a given directory hierarchy
# requires the xmllint command line tool which is part of the libxml2 library
for f in $(find . -type f -name '*.xml' | sort -V); do
echo "$f"; xmllint --format $f > foo && mv foo $f;
done
# find the ten EADs with the most lines
for f in $(find . -type f -name '*.xml'); do
wc -l $f
done | sort -nr | head
Sample Output:
281168 ./cbh/bcms_0002.xml
252915 ./tamwag/photos_223.xml
158820 ./tamwag/photos_023.xml
151900 ./akkasah/ad_mc_007.xml
127682 ./tamwag/tam_132.xml
114794 ./fales/mss_343.xml
112833 ./tamwag/tam_326.xml
109295 ./archives/rism_vf_001.xml
101747 ./fales/mss_208.xml
96752 ./tamwag/tam_415.xml
Takeaway:
Our largest EAD consists of 281,168 lines of XML.
Question: What is the maximum nesting level of each element?
Motivation:
As mentioned above, the EAD 2002 schema allows for certain elements to be nested arbitrarily deep. We needed to know how deep the element hierarchies were in practice in order to design the web pages. The element-depth-analyzer.rb script identifies the EADs with the deepest hierarchies for each element of interest. This was very useful for identifying candidate EADs used in development and testing, because the largest EAD does not necessarily have the deepest nesting level.
Script: element-depth-analyzer.rb
The element-depth-analyzer.rb is a simple Ruby script that relies on the Nokogiri Gem’s SAX stream parser. The script takes two arguments: the path to an EAD file to be analyzed, and the “element of interest” on which to run the depth analysis. The script simply responds to SAX parser events, incrementing a counter for each level of an element-of-interest hierarchy, (including the root element). The largest counter value is saved and output at the end of the script execution.
Sample Execution:
# Find the ten EADs with the deepest <c> element hierarchies
for f in $(find . -type f -name '*.xml'); do
bin/element-depth-analyzer.rb $f c
done | sort -nr | head
Sample Output:
# maximum depth observed, relative path to EAD
5,./vlp/mss_lapietra_001.xml
5,./tamwag/photos_097.xml
5,./tamwag/photos_019.xml
5,./nyhs/pr020_geographic_images.xml
5,./fales/mss_208.xml
5,./fales/mss_191.xml
5,./fales/mss_150.xml
5,./fales/mss_110.xml
5,./cbh/arms_2014_019_packer.xml
5,./cbh/arc_006_bergen.xml
Takeaway:
After running this script across the EAD corpus for each element of interest, we knew the depth of the nesting hierarchies that the design would need to accommodate, and we had identified EADs that would be useful for development and testing.
Does the EAD structure vary widely across archival repositories?
Motivation:
Although we believed that the data model was solid, we were concerned that there might be element attributes and parent-child element relationships in the EAD corpus that we did not anticipate. Therefore, we developed a pair of “element-union-analysis” scripts to gather this information.
Scripts: element-union-single.rb and element-union-multi.rb
The element-union-* scripts perform a recursive traversal of the parsed XML file(s). For each element type encountered, e.g., bioghist, dao, archdesc, a new “AnalysisNode” is created. The AnalysisNode is used to accumulate all of the attributes, child elements, nesting depth, and whether this element has siblings of the same type. All of the AnalysisNodes are stored in a hash keyed by the element name. During the traversal, the hash is queried to see if an AnalysisNode already exists for the element type. If not, a new AnalysisNode is created, otherwise the existing AnalysisNode is updated. At the end of the traversal, the scripts output all of the information in the AnalysisNodes, sorted by element name.
Two scripts were written to perform this analysis: the element-union-single.rb script is used to analyze individual EAD files, whereas the element-union-multi.rb script performs a union analysis across an arbitrary number of EADs. The script output can be loaded into a spreadsheet and compared against the data model. Additionally, the data from different runs can be loaded into a spreadsheet to compare the results from different sets of EADs. Please see below for an example.
Sample Execution:
element-union-single.rb fales/mss_208.xml element-union-multi.rb file-containing-paths-of-EADs-to-analyze.txt 1> results.txt
Sample Output Explanation:
# The line shown below # dao;true;0;["actuate", "href", "role", "show", "title", "type"];["daodesc"] # is interpreted as follows: # dao <-- the <dao> element... # true <-- has sibling <dao> elements... # 0 <-- is not nested inside other <dao> elements... # ["actuate", # "href", # "role", # "show", # "title", # "type"] <-- has the attributes listed inside the brackets, e.g., <dao actuate=... # ["daodesc"] <-- has child <daodesc> elements, i.e., <dao><daodesc>...</daodesc></dao>
Sample Output:
name;needs_array?;max_depth;attributes;children;child_sequences abstract;false;0;["id", "label"];["text"];["text"] accessrestrict;false;0;["id"];["head", "p"];["head_p"] appraisal;false;0;["id"];["head", "p"];["head_p"] archdesc;false;0;["level"];["accessrestrict", "appraisal", "arrangement", "bioghist", "controlaccess", "custodhist", "did", "dsc", "prefercite", "processinfo", "relatedmaterial", "scopecontent", "separatedmaterial", "userestrict"];["did_userestrict_accessrestrict_relatedmaterial_arrangement_scopecontent_bioghist_custodhist_prefercite_separatedmaterial_processinfo_appraisal_controlaccess_dsc"] arrangement;false;0;["id"];["head", "p"];["head_p+"] author;false;0;[];["text"];["text"] bioghist;false;0;["id"];["head", "p"];["head_p", "head_p+"] c;true;4;["id", "level", "otherlevel"];["bioghist", "c", "controlaccess", "did", "odd", "scopecontent", "separatedmaterial"];["did", "did_bioghist_scopecontent_c+", "did_bioghist_scopecontent_controlaccess_c+", "did_bioghist_scopecontent_separatedmaterial_controlaccess_c+", "did_c", "did_c+", "did_odd", "did_odd_scopecontent", "did_scopecontent", "did_scopecontent_c+", "did_scopecontent_controlaccess_c+", "did_separatedmaterial"] change;false;0;[];["date", "item"];["date_item"] container;true;0;["altrender", "id", "label", "parent", "type"];["text"];["text"] controlaccess;false;0;[];["corpname", "genreform", "geogname", "occupation", "persname", "subject"];["genreform+_geogname", "genreform+_geogname+", "genreform+_geogname+_genreform+_geogname_genreform+_geogname", "genreform+_geogname_genreform+_geogname_genreform+", "genreform+_geogname_genreform+_geogname_genreform+_geogname_genreform+_subject_genreform_geogname_genreform+_subject+", "genreform+_persname", "genreform+_subject+_geogname+_genreform_subject_genreform+_geogname+_genreform_geogname+_subject_geogname+", "genreform_subject+_genreform_geogname_genreform+_subject_geogname+_subject+_geogname+_subject+_geogname_subject_geogname_subject_genreform+_subject_genreform+_subject_genreform_subject+_genreform_subject_occupation_subject_genreform+_subject+_genreform+_subject+_genreform+_geogname_genreform+_geognam\ e_genreform_geogname+_corpname_persname+_corpname_persname+_corpname_persname_corpname", "genreform_subject+_geogname_subject_genreform+_geogname_genreform+_persname", "geogname+_genreform_geogname+_genreform_geogname_genreform+_geogname_genreform+_persname", "geogname+_persname_corpname", "geogname_genreform+_subject_genreform+_geogname_subject_geogname+_genreform+_persname", "occupation_geogname_genreform+_subject_genreform+_subject+_geogname_genreform_subject_geo\ gname_persname+_corpname"] … p;true;0;[];["date", "emph", "lb", "text", "title"];["date", "emph", "emph_text", "emph_text_emph_text", "emph_text_emph_text_emph_text", "emph_text_emph_text_emph_text_emph_text", "emph_text_emph_text_emph_text_emph_text_emph_text", "text", "text_emph_text", "text_emph_text_emph_text", "text_lb+_text\ ", "title_text"] persname;true;0;["role", "rules", "source"];["text"];["text"] physdesc;false;0;["altrender", "id", "label"];["extent"];["extent", "extent+"] prefercite;false;0;["id"];["head", "p"];["head_p"] processinfo;false;0;["id"];["head", "p"];["head_p+"] profiledesc;false;0;[];["creation", "langusage"];["creation_langusage"] publicationstmt;false;0;[];["p", "publisher"];["publisher_p"] publisher;false;0;[];["text"];["text"] relatedmaterial;false;0;["id"];["head", "p"];["head_p+"] repository;false;0;[];["corpname"];["corpname"] revisiondesc;false;0;[];["change"];["change"] scopecontent;false;0;["id"];["head", "p"];["head_p", "head_p+"] separatedmaterial;false;0;["id"];["head", "p"];["head_p", "head_p+"] sponsor;false;0;[];["text"];["text"] subject;true;0;["source"];["text"];["text"] text;false;0;[];[];[""] title;false;0;["render"];["text"];["text"] titleproper;false;0;[];["num", "text"];["text_num"] titlestmt;false;0;[];["author", "sponsor", "titleproper"];["titleproper_author_sponsor"] unitdate;false;0;["datechar", "normal", "type"];["text"];["text"] unitid;false;0;[];["text"];["text"] unittitle;false;0;[];["emph", "text", "title"];["emph", "emph_text", "emph_text_emph", "text", "text_emph", "text_emph_text", "title_text"] userestrict;false;0;["id"];["head", "p"];["head_p"]
Sample Analysis Spreadsheet:
Over the course of the FADESIGN project we kept increasing the size of the EAD corpus used for development and testing. Sometimes the additional EADs were from an organization for whom we already had EADs, while at other times we received EADs from an organization that we hadn’t sampled before. We would run the element-union-* scripts on these sample sets and load the results into a spreadsheet for analysis.
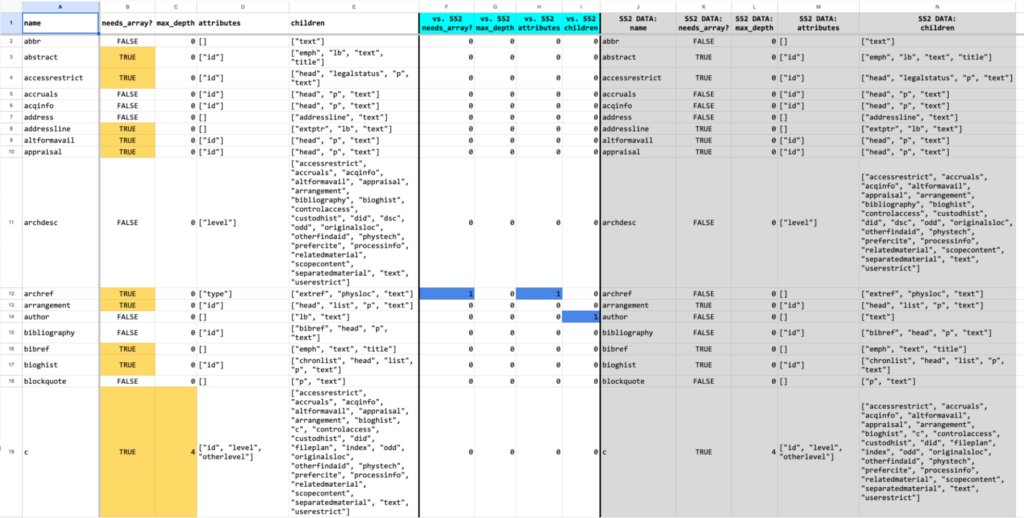
In the spreadsheet excerpt below, the results of two sample sets from the same organization are being compared: Sample Set 3 vs. Sample Set 2 (SS2). As you can see in columns F, H, and I, there are differences for the <archref> and <author> elements. This information was used to ensure that the data model and corresponding software data structures accommodated these use cases.
Spreadsheet excerpt showing the comparison of data from two EAD sample sets:
Takeaway:
The data collected with these scripts was enormously useful. It not only helped us refine the data model and software data structures as our EAD corpus grew, but also allowed us to compare the EAD structure across archival repositories.
Question: How many times does an element appear in the EAD?
Motivation:
We wanted to identify EADs that would be suitable candidates for various testing scenarios.
Script: gen-element-counts.sh
The gen-element-counts.sh script, written in Bash, contains a simple set of commands that leverage the grep utility’s “extended regular expression” and “count” functionality. The script prints the results in comma separated value (CSV) format to STDOUT. By redirecting the script output to a .csv file, one can import and review the results using a spreadsheet application. Sorting the spreadsheet by column in descending order, one can see which elements appear most frequently in the EAD.
Examples are shown below.
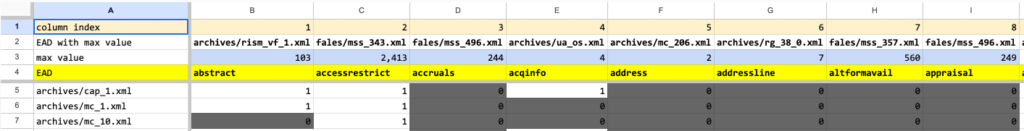
One may ask why the data isn’t column oriented instead of row oriented,e.g., “Shouldn’t column A contain the element names and column B contain the element counts?” The row-oriented output allows one to run the script over an arbitrary number of EADs, directing the output to a file. The file can be processed (see below), and the results loaded into a spreadsheet application. This allows one to determine the EADs with the largest number of a given element across the entire input set. The third spreadsheet excerpt below demonstrates this use case, e.g., fales/mss_343.xml has the greatest number of accessrestrict elements at 2,343.
Spreadsheet excerpt for a single EAD after importing the csv file:
Spreadsheet excerpt for a single EAD after sorting columns B through CR in descending order by row 2
Spreadsheet excerpt containing data from 2,249 EADs. The spreadsheet contains a formula that identifies the EAD with the greatest number of instances for a given element.
Sample Execution:
gen-element-counts.sh ./tamwag/photos_223.xml > tamwag-photos_223-element_counts.csv
Sample Output:
FILE,abstract,accessrestrict,accruals,acqinfo,address,addressline,altformavail,appraisal,archdesc,archref,arrangement,bibliography,bibref,bioghist,blockquote,c,change,chronitem,chronlist,colspec,container,controlaccess,corpname,creation,custodhist,dao,daodesc,daogrp,daoloc,date,defitem,did,dimensions,dsc,eadheader,eadid,editionstmt,emph,entry,event,eventgrp,extent,extptr,extref,famname,filedesc,fileplan,function,genreform,geogname,head,index,indexentry,item,langmaterial,language,langusage,legalstatus,list,materialspec,name,note,notestmt,num,occupation,odd,originalsloc,origination,otherfindaid,p,persname,physdesc,physfacet,physloc,phystech,prefercite,processinfo,profiledesc,publicationstmt,relatedmaterial,repository,revisiondesc,row,scopecontent,separatedmaterial,subject,table,tbody,tgroup,title,titleproper,titlestmt,unitdate,unittitle,userestrict,
tamwag/photos_223.xml,1,1,0,1,1,7,0,0,1,0,2,1,4,1,0,26504,2,0,0,0,53005,2,313,1,0,0,0,0,0,4,0,26505,0,1,1,1,0,117,0,0,0,2,1,0,0,1,0,0,3,5,3720,3700,9907,2,1,0,1,0,0,0,6420,0,0,1,0,1,0,3,0,63,76,1,0,0,1,1,2,1,1,2,1,1,0,5,1,3204,0,0,0,2,1,1,26248,26505,1,
Sample Data Processing:
# ----------------------------------------------------------
# extract-element-count script execution / data processing
# ----------------------------------------------------------
# extract element counts
time for f in $(cat file-list.txt); do ./bin/gen-element-counts.sh $f | tee -a element-counts.txt; done
# extract header line
head -1 element-counts.txt > element-counts.csv
# extract data and append to csv file
grep -v FILE element-counts.txt >> element-counts.csv
# check that the line count is the EAD file count + 1 (for the header row)
wc -l element-counts.csv
2112 element-counts.csv
Takeaway:
It is useful to know which EAD in a sample set has the maximum occurrences of a given element. The data can be used to identify test candidates and also provides data useful for correlating software performance against EAD element counts . For example, during our EAD indexing we repeatedly observed that certain EADs took significantly longer to index than others. We found that these EADs had the highest number of container (<container>) elements. We optimized the code for container element processing which reduced the application execution time by orders of magnitude.
Question: Across all EADs, what sequences of child elements are present?
Motivation:
As we worked on this project, we realized one problem that is actually noted in the Golang documentation:
"Mapping between XML elements and data structures is inherently flawed: an XML element is an order-dependent collection of anonymous values, while a data structure is an order-independent collection of named values."
Development was already quite far along when we realized this. Instead of changing our entire parsing strategy, which would have required significant rework of our Hugo templates, we decided to selectively implement stream parsing to preserve the element order when necessary . To determine which elements needed to be stream parsed, we created scripts that were variations of the element-union-* scripts described above. These new scripts output the same data as the element-union-* strings, but add child-element sequence information that we used to determine where stream parsing was required.
Scripts: element-union-with-child-sequences-single.rb and element-union-with-child-sequences-multi.rb
Sample Execution:
element-union-with-child-sequences-single.rb ./fales/mss_208.xml
element-union-with-child-sequences-multi.rb file-containing-paths-of-EADs-to-analyze.txt 1> results.txt
Sample Output Explanation:
# In the output below, a child sequence containing a "<element name>+" # indicates that there was more than one consecutive element of that type. # # For example, the line # bioghist;false;0;["id"];["head", "p"];["head_p", "head_p+"] # can be broken down as follows: # bioghist; <-- element name # false; <-- does not have any sibling <bioghist> elements # 0; <-- is not nested inside any other <bioghist> elements # ["id"]; <-- has an @id attribute # ["head", "p"]; <-- has the child elements <head> and <p> # ["head_p", "head_p+"] <-- the <bioghist> has child elements in # the following sequences: # <head></head><p></p> # and # <head></head><p></p><p></p>...
Sample Output:
name;needs_array?;max_depth;attributes;children;child_sequences abstract;false;0;["id", "label"];["text"];["text"] accessrestrict;false;0;["id"];["head", "p"];["head_p"] appraisal;false;0;["id"];["head", "p"];["head_p"] archdesc;false;0;["level"];["accessrestrict", "appraisal", "arrangement", "bioghist", "controlaccess", "custodhist", "did", "dsc", "prefercite", "processinfo", "relatedmaterial", "scopecontent", "separatedmaterial", "userestrict"];["did_userestrict_accessrestrict_relatedmaterial_arrangement_scopecontent_bioghist_custodhist_prefercite_separatedmaterial_processinfo_appraisal_controlaccess_dsc"] arrangement;false;0;["id"];["head", "p"];["head_p+"] author;false;0;[];["text"];["text"] bioghist;false;0;["id"];["head", "p"];["head_p", "head_p+"] c;true;4;["id", "level", "otherlevel"];["bioghist", "c", "controlaccess", "did", "odd", "scopecontent", "separatedmaterial"];["did", "did_bioghist_scopecontent_c+", "did_bioghist_scopecontent_controlaccess_c+", "did_bioghist_scopecontent_separatedmaterial_controlaccess_c+", "did_c", "did_c+", "did_odd", "did_odd_scopecontent", "did_scopecontent", "did_scopecontent_c+", "did_scopecontent_controlaccess_c+", "did_separatedmaterial"] change;false;0;[];["date", "item"];["date_item"] container;true;0;["altrender", "id", "label", "parent", "type"];["text"];["text"] controlaccess;false;0;[];["corpname", "genreform", "geogname", "occupation", "persname", "subject"];["genreform+_geogname", "genreform+_geogname+", "genreform+_geogname+_genreform+_geogname_genreform+_geogname", "genreform+_geogname_genreform+_geogname_genreform+", "genreform+_geogname_genreform+_geogname_genreform+_geogname_genreform+_subject_genreform_geogname_genreform+_subject+", "genreform+_persname", "genreform+_subject+_geogname+_genreform_subject_genreform+_geogname+_genreform_geogname+_subject_geogname+", "genreform_subject+_genreform_geogname_genreform+_subject_geogname+_subject+_geogname+_subject+_geogname_subject_geogname_subject_genreform+_subject_genreform+_subject_genreform_subject+_genreform_subject_occupation_subject_genreform+_subject+_genreform+_subject+_genreform+_geogname_genreform+_geognam\ e_genreform_geogname+_corpname_persname+_corpname_persname+_corpname_persname_corpname", "genreform_subject+_geogname_subject_genreform+_geogname_genreform+_persname", "geogname+_genreform_geogname+_genreform_geogname_genreform+_geogname_genreform+_persname", "geogname+_persname_corpname", "geogname_genreform+_subject_genreform+_geogname_subject_geogname+_genreform+_persname", "occupation_geogname_genreform+_subject_genreform+_subject+_geogname_genreform_subject_geo\ gname_persname+_corpname"] … p;true;0;[];["date", "emph", "lb", "text", "title"];["date", "emph", "emph_text", "emph_text_emph_text", "emph_text_emph_text_emph_text", "emph_text_emph_text_emph_text_emph_text", "emph_text_emph_text_emph_text_emph_text_emph_text", "text", "text_emph_text", "text_emph_text_emph_text", "text_lb+_text\ ", "title_text"] persname;true;0;["role", "rules", "source"];["text"];["text"] physdesc;false;0;["altrender", "id", "label"];["extent"];["extent", "extent+"] prefercite;false;0;["id"];["head", "p"];["head_p"] processinfo;false;0;["id"];["head", "p"];["head_p+"] profiledesc;false;0;[];["creation", "langusage"];["creation_langusage"] publicationstmt;false;0;[];["p", "publisher"];["publisher_p"] publisher;false;0;[];["text"];["text"] relatedmaterial;false;0;["id"];["head", "p"];["head_p+"] repository;false;0;[];["corpname"];["corpname"] revisiondesc;false;0;[];["change"];["change"] scopecontent;false;0;["id"];["head", "p"];["head_p", "head_p+"] separatedmaterial;false;0;["id"];["head", "p"];["head_p", "head_p+"] sponsor;false;0;[];["text"];["text"] subject;true;0;["source"];["text"];["text"] text;false;0;[];[];[""] title;false;0;["render"];["text"];["text"] titleproper;false;0;[];["num", "text"];["text_num"] titlestmt;false;0;[];["author", "sponsor", "titleproper"];["titleproper_author_sponsor"] unitdate;false;0;["datechar", "normal", "type"];["text"];["text"] unitid;false;0;[];["text"];["text"] unittitle;false;0;[];["emph", "text", "title"];["emph", "emph_text", "emph_text_emph", "text", "text_emph", "text_emph_text", "title_text"] userestrict;false;0;["id"];["head", "p"];["head_p"]
Takeaway:
It was critical to understand the child element sequences so that we could ensure that our code generated HTML that reflected the original order found in the EADs.
Questions:
What is the set of <dao @xlink:role> values across all EADs?
Are there EADs with <dao @xlink:role> values outside of the controlled vocabulary?
Motivation:
Our finding aids allow patrons to directly access various types of digital content, e.g., audio, video, images, or request access to other content, e.g., electronic records. The finding aids publishing infrastructure needs to know what type of digital content is being served so that the appropriate viewer can be included in the HTML, e.g., an image view, audio player, video player. The EAD Digital Archival Object (DAO) elements have role attributes that indicate what kind of digital object is being described, e.g., image-service, audio-service, video-service, electronic-records-reading-room. These role attribute values come from a controlled vocabulary. In order to ensure that our controlled vocabulary contained all valid DAO role values in the EADs, and to identify EADs with DAO roles that required remediation, we wrote a script that extracts all of the unique <dao @xlink:role> values across a set of EADs.
Script: dao-role-extractor-multi.rb
Sample Execution:
dao-role-extractor-multi.rb file-containing-paths-of-EADs.txt 1> results.txt
Sample Output:
processing tamwag/aia_001.xml
processing tamwag/aia_002.xml
processing tamwag/aia_003.xml
processing tamwag/aia_004.xml
processing tamwag/aia_005.xml
processing tamwag/aia_006.xml
processing tamwag/aia_007.xml
processing tamwag/aia_008.xml
processing tamwag/aia_009.xml
processing tamwag/aia_010.xml
…
processing tamwag/wag_370.xml
processing tamwag/wag_371.xml
processing tamwag/wag_372.xml
processing tamwag/wag_373.xml
processing tamwag/wag_375.xml
processing tamwag/web_arc_001.xml
processing tamwag/web_arc_002.xml
processing tamwag/web_arc_003.xml
# the contents of the results.txt is as follows:
ROLES:
audio-reading-room
audio-service
electronic-records-reading-room
external-link
image-service
video-reading-room
video-service
Takeaway:
Being able to extract the DAO roles across an arbitrary set of EADs helped us identify non-conforming EADs and finalize our <dao @xlink:role> controlled vocabulary.
Acknowledgements:
Thanks to Laura Henze, Deb Verhoff, and Don Mennerich for reviewing this post and providing feedback.